- support
- info@evidentic.com
- +49 (0) 30 959 99 8831
1A

3B026B

NDS3PML

378984

G023GS

1197738

AHVE13A

KT0KBL2

1996533

1979430
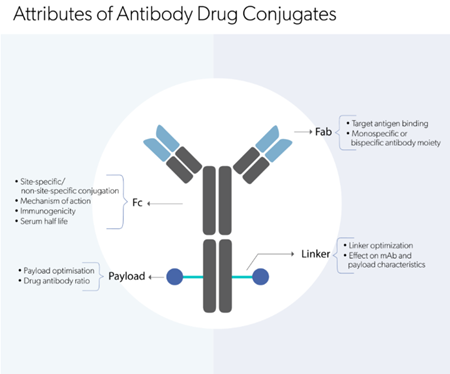
Explore our comprehensive suite of analytical
methods to analyze biologic molecules.










Hilton Amsterdam Airport Schiphol
Schiphol Boulevard 701, 1118 BN Schiphol/Netherlands
Omni Boston Hotel at the Seaport
450 Summer Street, Boston, MA 02210/US
Evidentic GmbH
Martin-Buber-Str. 10
14163 Berlin